
The Human Genome Project concluded with the successful sequencing of the first human genome in 2003. Since then medical breakthroughs enabled by unlocking the secrets of our DNA have been much touted, yet hardly delivered. Complete Genomics, a world leader of complete genome sequencing, wants to change that. Singularity Hub sat down with co-founder and CEO, Cliff Reid, and asked him about a future of genetics-based health care and the role Complete Genomics will play in ushering in this new era of medicine.
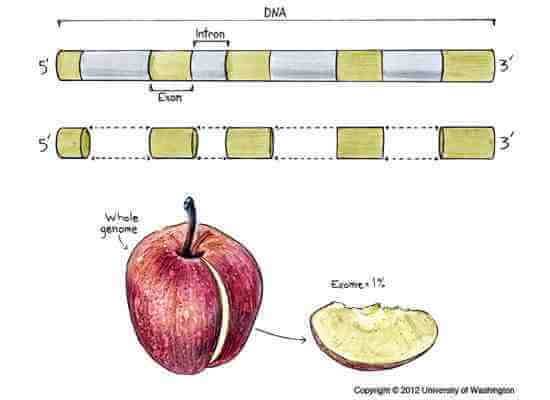
According to their website, Complete Genomics is the first company dedicated solely to sequencing whole human genomes. While other companies offer outsourced sequencing, many of these services sequence the ‘exome,’ the stretches of DNA that code for proteins and make up just one percent of the human genome, or specialize in sequencing the genomes of mice, rats, and other animals used in research. Focusing on whole human genome sequencing, Reid insists, gives Complete Genomics the edge in the race to truly understand the human genome in its entirety.
That’s a tall order to say the least, but the company is taking a new direction that could steepen the learning curve. In this exclusive one on one interview, Singularity Hub’s Aaron Saenz speaks with Complete Genomics CEO Cliff Reid about where Complete Genomics is now, and where they are going:
Until now all of their customers have been researchers. Major institutions like the National Cancer Institute and the Northern Virginia Healthcare system (INOVA) employ Complete Genomes’ services to conduct their research. But by the end of the year, Reid says, their whole genome sequencing will be offered to the clinical sector. Patients’ genomes will be sequenced and be made available to their doctors. Reid thinks that the clinic is where the true future of complete genome sequencing lies: he expects the clinical sequencing market to far exceed the size of the research market.
The company will be aiming its efforts at two specific clinical areas: idiopathic children, or children who are ill yet undiagnosed for a disease, and cancer. Idiopathic children often remain undiagnosed for years, moving from pediatrician to pediatrician. By screening their entire genomes for all known diseases, the company hopes to help doctors turn mysteries into diagnoses.
The second market, Reid thinks, will be even bigger. Cancer is a disease of the genome. As cancer researchers find out more about the genetic mistakes that cause tumors, eventually the sequences of all tumors will be known.
Not all tumor cells are alike. As they grow the cells mutate in different ways. One thing Complete Genomics is currently working on is technology that would allow them to sequence the genomes of single cells. Sequencing a group of tumor cells they could compare the different cells and watch how their individual mutations progress over time. The changes would allow them to “run the tumor movie.” Or, as Reid argues, “most importantly run the tumor movie backwards, because when you run it back to zero time you find the mutation that actually caused that tumor.”
Reid expects single cell sequencing to be commercially available within two years, and he’s very optimistic about the potential of whole genome sequencing in the fight against cancer. “I don’t think my kids are going to worry about cancer. I think we’re going to nail it in my lifetime. We’re never going to be able to stamp it out [completely] because they are mutations, and mutations are going to happen. But we’re going to be able to treat it. We’re going to turn cancer into a chronic disease, not a death sentence.”
But sequencing is only half the battle.
Reading the three billion base pairs is not only an enormous task but an incredibly complex one. The speed of next generation sequencing has created a gap between data generation and data analysis. Making analysis more user-friendly is something Reid thinks will emerge with developing technologies. “One of the things going on is the wholesale growth of the informatics community, the analysis community, so that they can take these stunningly rich data sets that we’re producing now and really get all of the information out of them. Not just statistical correlations but real understanding of the biological information.”
Yet, he admits the field still has a very long way to go. “When will that promise actually be realized? I think that for us to understand the genetic basis of human disease in all of its complexity – we’ll never understand all of it, but to get most of it – I think we need to sequence about a million genomes. To date we’ve sequenced about ten thousand. We’re a solid one percent of the way there.”
From Reid’s perspective, the company’s just getting started.

Complete Genomics has benefited from next generation sequencing technologies that have halved their cost per genome each year for three years straight. Their current capacity is 10,000 genomes per year and they’re read with stunning accuracy. In July they published a study in Nature demonstrating a new sequencing method that makes just one error per 10 million base pairs. To put that in perspective, such accuracy yields a minuscule 600 errors across the entire 3 million base pairs of the human genome. Practically speaking, it’s errorless.
That accuracy was published in a study in the Aug. 3 issue of Cell. Sarah Tishkoff’s team at the University of Pennsylvania used Complete Genomics’ whole genome sequencing to explore the genetic diversity of human origins. Comparing the genomes of three separate African populations the researchers discovered 13.4 million new genetic variations. Only 72,000 of these variations were found in the coding region, the rest in the long stretches of DNA between genes that don’t code for genes but control how genes are expressed. The multitude of variants found in the non-coding region underscores the importance of sequencing the whole genome and not merely the exome if we’re ever to understand the full complexity of our genetic underpinnings. The study may also have unveiled two exciting discoveries. In each of the three groups they identified sequences that appear to have originated from a hominin lineage other than Homo sapiens. The researchers speculate that it could be a species closely related to Neanderthal. Additionally, one of the populations studied was the Cameroon Pygmies. The study identified several genes involved with pituitary gland function, which affects growth, that may explain why Pygmies are short.
Trying to stay ahead of the exponential sequencing curve, Complete Genomics engineers are already designing the next, next generation technologies. They’ll not only be virtually errorless but also much faster. When their fleet of new sequencers come online the company’s sequencing capacity will increase to one million whole genomes per year.
Reid’s vision of individualized, genetics-based medicine draws closer as the cost of sequencing continues to drop and the rate of sequencing continues to climb. He is one hundred percent sure that there will come a day when our whole genome sequence will be an integral part of our everyday healthcare.
“Inevitably there’s going to be a tipping point where whole human genome sequencing becomes ubiquitous. I think that tipping point will be driven by more than price. I think it will be driven by a combination of three things: one is price, the second is utility – how valuable is it for you to understand what’s in your genome. And then the third is a general acceptability, kind of a network effect of physicians being able to speak about it, your friend down the street…also having his or her genome sequenced so you can talk about it. There’s this critical mass effect that hasn’t yet happened in complete human genome sequencing and I expect it will happen in the next few years. I don’t think price alone will tip it. I think the research community must continue to learn more and more about the genome to make it more valuable for you and me to have our genome sequenced as part of our routine healthcare.”
[image credits: University of North Texas, University of Washington, and Xconomy]
images: University of North Texas, University of Washington, and Xconomy
video: Singularity University via YouTube


