You know those stories of scientific breakthroughs, in which the lone genius scientist struggles for years until his “eureka!” moment?
Yeah, that’s a lie.
With the big data revolution well under way, today scientific discoveries are the result of massive collaborations.
Case in point? Last week, 18 institutions teamed up and devised a method to image entire brains 1,000 times faster than anything before. Dubbed by the team as an “Avengers, unite!” moment, they combined strengths to physically blow up brain tissue to over 20 times its usual size, and scanned their inner circuits and molecular constituents—down to the nano-scale level—using a new type of blazingly fast microscopy.
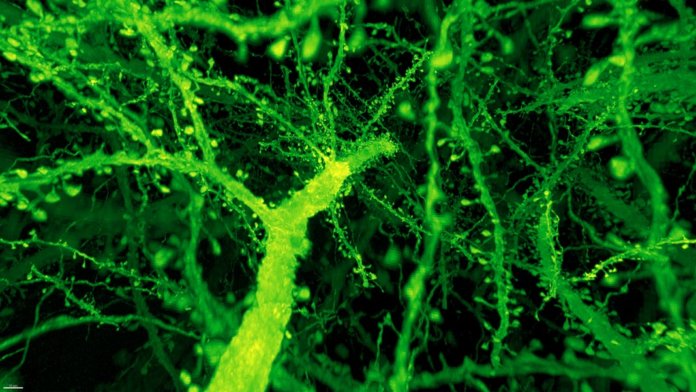
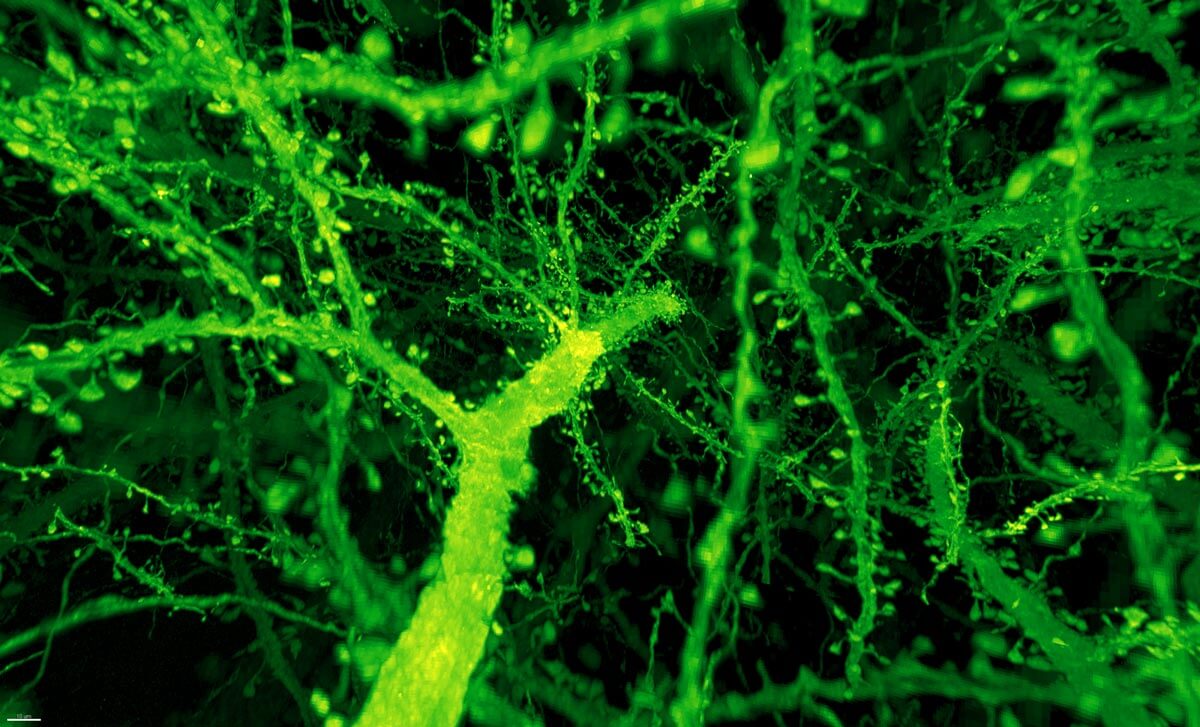
The results are stunning videos of the fly and mouse brains, in which every nook and cranny is illuminated and reconstructed in neon colors.
It’s not just eye-candy. Scientists have been eagerly devising new ways to map entire brains with increasing precision and resolution, with the hope of unlocking the brain’s mysteries—which circuits underlie what behaviors? How are memories formed, saved, and retrieved? How do circuits reorganize with learning or age?
But the end game is vastly more ambitious: simulate a whole brain inside a computer, a feat that some say will eventually lead to general AI. Previous attempts were stymied by the years it took to painstakingly piece together a single brain.
But now, “we’ve crossed a threshold in imaging performance,” said Dr. Ed Boyden at MIT, one of the leading investigators in this project. “That’s why we’re so excited. We’re not just scanning incrementally more brain tissue, we’re scanning entire brains.”
The teams published their results in Science.
The Brain Imaging Cheat Sheet
Most of us have heard of MRI as a way to image the brain and suss out potential problems in the clinic.
But things get much weirder in the lab, where scientists are mapping out every twist and turn of a neuron’s branches and their connections—synapses, which mainly sit on little protrusions on a branch called a dendritic spine.
To tease apart structures within the fatty mush of tissue, scientists have long used tiny little protein lights to illuminate parts of the brain. By tinkering with an animal’s genome, scientists can “tag” these lights—in all colors of the rainbow—to specific structures such as the synapses or the branches.
They then painstakingly take out the brain, slice it into wafer-thin sections, and treat those sections in various ways to enhance the light’s brilliance under the microscope. The sections are imaged one by one, and stitched back together into 3D using advanced algorithms.
It’s a painfully tedious process that takes weeks, if not months or years. What’s more, because microscopes are inherently limited in resolution, the reconstructions can end up blurry—not a great place to start if you want to simulate a brain.
Alice in Wonderland
A few years back, Boyden tackled the problem with a seemingly nutty solution: why not physically expand the brain, so that it’s details are easier to see?
Using a swellable gel found in—not joking—diapers, Boyden’s team encapsulated brains inside the material and upon adding water, blew the brains up to roughly 20 times their original size.
You’d think the process would tear delicate brain tissue apart. Instead, the neurons and circuits were protected by the gel, and every single molecule within the brain kept its relative position after expansion.
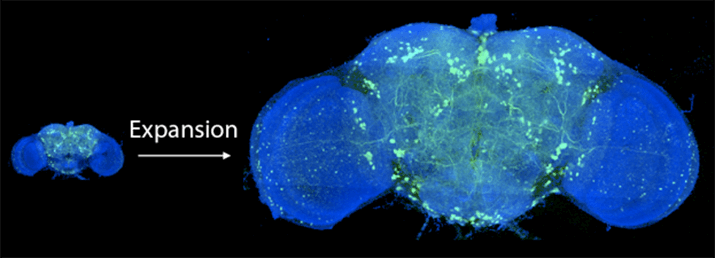
Scientists around the world have since adapted the technique, resulting in stunning findings about our brain’s organizational patterns. Yet the teams all ran into a problem: with greater scale came greater imaging times, which made mapping large circuits a chore and whole brains impossible. What’s more, the longer you zap the light proteins, the easier it is to bleach the protein—resulting in black spots in the image—or photodamage the brain tissue.
Large-Scale Imaging
That’s when Boyden gave Dr. Eric Betzig at HHMI’s Janelia Research Campus a call.
Back in 2014, Betzig and colleagues developed a powerful new tool that rapidly collects high-resolution images and minimizes damage. The technology, “lattice light-sheet microscope,” sweeps a thin sheet of light multiple times across every plane of the brain tissue. This keeps only one focal plane in view, which helps minimize blurriness.
The scope also splits up its light beams in a way that greatly reduces the intensity of the light—the lower the intensity, the less photodamage, which allows researchers to image the brain for longer periods without frying their precious samples.
What’s more, because the light captures a whole plain instead of a set of points, it captures images much faster, without sacrificing resolution.
The microscope allowed Betzig and team to track the subcellular dynamics of healthy, living cells as they went about their business. The results caught Boyden’s eye: expansion microscopy, which turns brains virtually translucent, could be perfect for Betzig’s technology, which shines light from one side and snaps a photo from the other.
Betzig was more cynical.
“I thought they were full of it…I was going to show them,” he said.
Instead, the combo proved much more powerful than anyone expected. Under Betzig’s microscope, expanded mouse tissue gave up all its structural secrets. Dotted along each neuron’s branches were mushroom-shaped protrusions, dendritic spines, that are normally hard to see clearly—the “neck” of the mushroom often looks like a blurry smudge. Instead, even the smallest necks were in sharp focus.
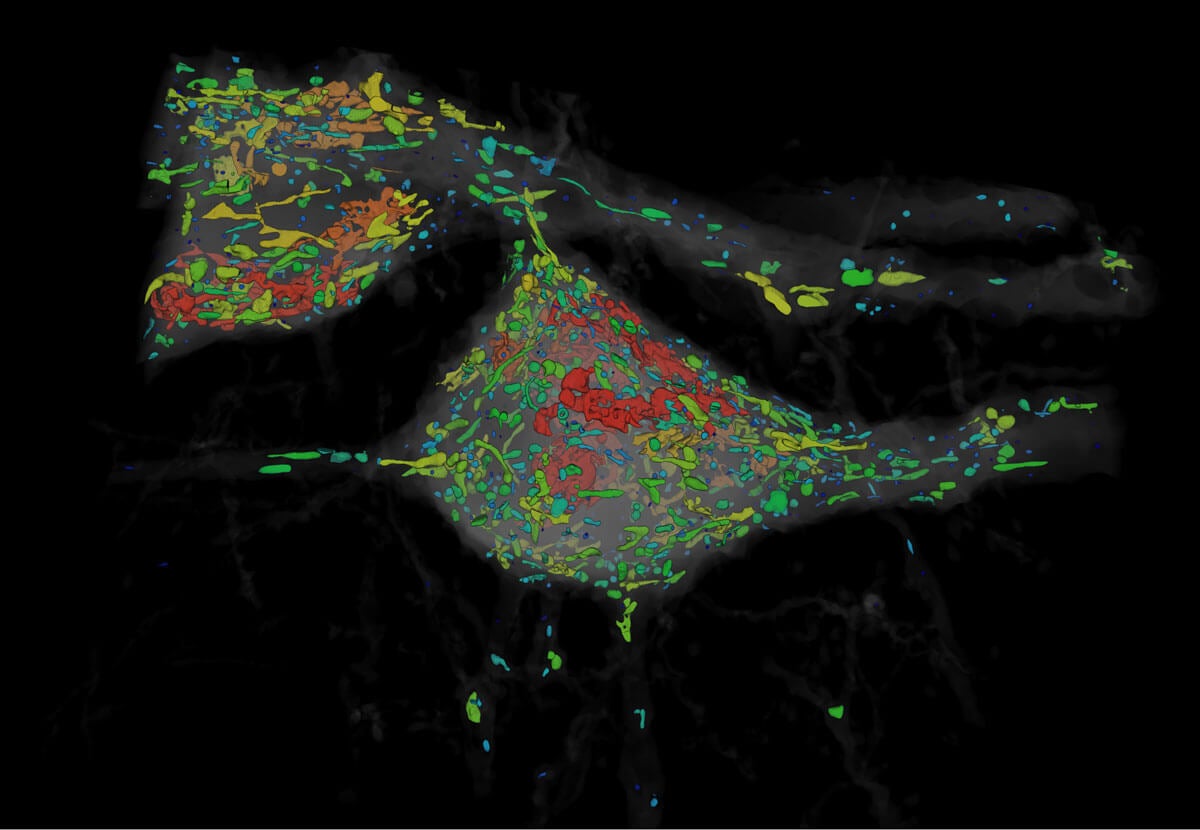
The team then compared the density of synapses in various parts of the mouse’s cortex as proof-of-concept, and the new technique allowed them to analyze millions of synapses in just a few days.
“Using electron microscopy, this would have taken years to complete,” said first author Dr. Ruixuan Gao.
Whole-Brain Revolution
Encouraged, the team turned to whole brains, starting with the fly. A fly’s brain is roughly the size of a poppy seed and contains about 100,000 neurons linked up in complex circuits.
Breaking down the brains into 50,000 “cubes” of 3D blocks, the team devised new algorithms that stitched the blocks back together like a puzzle. Other members then came in to wrangle over 40 million synapses into stunning, neon-colored visuals to make the data more interpretable.
In one set of experiments, the team traced a smell-related circuit across several brain regions, identified a specific set of neurons within those circuits, and counted all of their synapses. It’s a scale and depth previously unimaginable.
Nevertheless, the whole-brain imaging revolution is just starting, and the team believes there’s more to improve. Certain brain regions don’t like to be tagged with protein light bulbs, and it’s always hard to squeeze enough in to light up tiny areas. Not all tissues like to be stretched—for example, collagen, the stuff that makes up connective tissue. Perhaps most importantly, each processing step could introduce artifacts into the tissue, which means scientists will have to carefully validate their results.
But the future looks brighter than ever before. Scientists could finally look at large-scale maps of the brain to pinpoint changes that drive brain disorders, explain how we make memories and decisions, or track circuit changes throughout an entire lifetime. In time, we could get to maps of entire nervous systems.
“That’s like the holy grail for neuroscience,” said Boyden.
Image Credit: Image from Gao et al., SCIENCE Vol. 363, Issue 6424, eaau8302 (18 Jan 2019). Republished with permission from American Association for the Advancement of Science (AAAS).