This Radical New DNA Microscope Reimagines the Cellular World
Share
It’s not every day that something from the 17th century gets radically reinvented.
But this month, a team from the Broad Institute at MIT and Harvard took aim at one of the most iconic pieces of lab ware—the microscope—and tore down the entire concept to recreate it from scratch.
I’m sure you have a mental picture of a scope: a stage to put samples on, a bunch of dials to focus the image, tunnel-like objectives with optical bits, an eyepiece to observe the final blown-up image. Got it? Now erase all that from your mind.
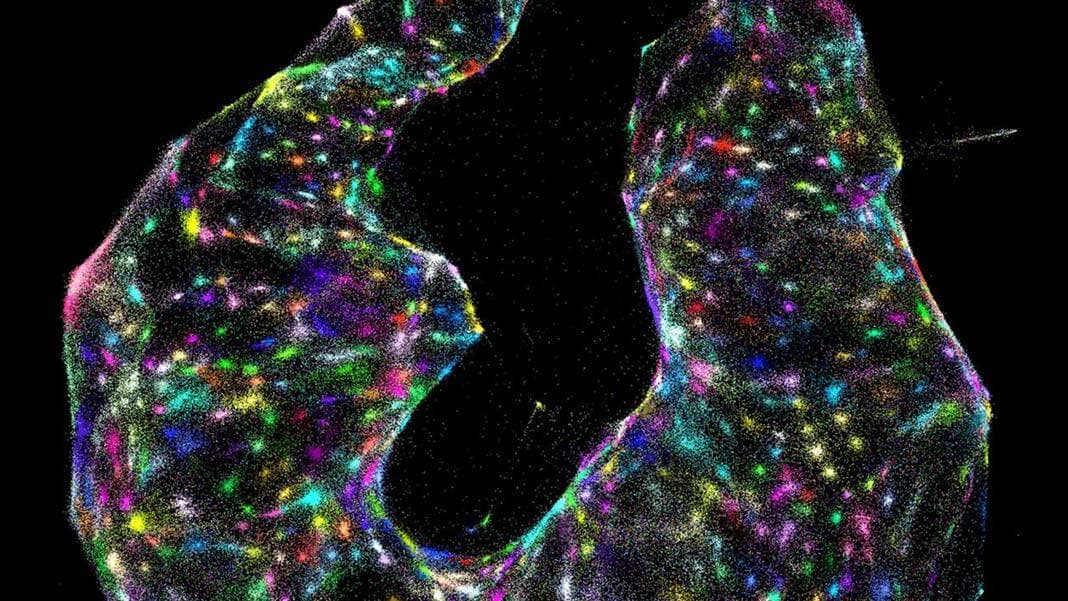
The new technique, dubbed DNA microscopy, uses only a pipette and some liquid reagents. Rather than monitoring photons, here the team relies on “bar codes” that chemically tag onto biomolecules. Like cell phone towers, the tags amplify, broadcasting their signals outward. An algorithm can then piece together the captured location data and transform those GPS-like digits into rainbow-colored photos.
The results are absolutely breathtaking. Cells shine like stars in a nebula, each pseudo-colored according to their genomic profiles.
That’s the crux: DNA microscopy isn’t here to replace its classic optical big brother. Rather, it pulls the viewer down to a molecular scale, allowing you to see things from the “eyes of the cell,” said study author Dr. Joshua Weinstein, who worked under the supervision of Dr. Aviv Regev and Dr. Feng Zhang, both Howard Hughes Medical Institute investigators.
The tech decodes the natural location of molecules inside a cell—and how they interact—while simultaneously gathering information about its gene expression in a two-in-one combo. It’s a bonanza for scientists struggling to tease apart individual differences in cells that physically look identical—say, immune cells that secrete different antibodies, or cancer cells in their early stage of malignant transformation.
“It’s a completely new way of visualizing biology,” said Weinstein in a press release. “This gives us another layer of biology that we haven’t been able to see.”
Why?
Almost all current microscopy techniques stem from the original all-in-one light microscope, first introduced in the 17th century. The core concept is light: the device guides photons refracted from the sample into a series of mirrors and optical lenses, resulting in an enlarged image of whatever you’re focusing on. It basically works like a DSLR camera with a very powerful zoom lens.
Scientists have long since moved past the visible light spectrum. Electron microscopy, for example, measures electrons that bounce off tissue to look at components inside the cell. Fluorescent microscopy—the “crown prince” of imaging—captures emitted light waves after stimulating tissue-bound fluorescent probes with lasers.
But here’s the thing: even as traditional microscopy is increasingly perfected and streamlined, it hits two hard limits. One is resolution. Light scatters, and there’s only so much we can do to focus the beam on one point to generate a clear image. This is why a light microscope can’t clearly see DNA molecules or proteins—it’s like using a smartphone to capture a single bright star. As a result, current microscopes generate satellite views of goings-on on the cellular “Earth.” Sharp, but from afar.
The second is genomic profiling. There’s been a revolution in mapping cellular diversity to uncover how visually similar cells can harbor dramatically different genomic profiles. A perfect example is the immune system. Immune cells that look similar can secrete vastly different antibodies, or generate different protein “arms” on their surface to grab onto specific types of cancer cells. Sequencing the cells’ DNA loses spatial information, making it hard to tease out whether location is important for designing treatments.
So far, microscopy has only focused on half of the picture. With DNA microscopy, the team hopes to grab the entire landscape.
“It will allow us to see how genetically unique cells—those comprising the immune system, cancer, or the gut, for instance—interact with one another and give rise to complex multicellular life,” explained Weinstein.
Inner Workings
To build their chemical microscope, the team began with a group of cultured cells.
They decoded the cells’ RNA molecules and transcribed the data to generate a complete library of expressed genes called cDNA. Based on cDNA sequences, they then synthesized a handful of tags randomly made of the DNA letters A, T, C, and G, each about 30 letters long. When bathed onto a new batch of cells, the tags tunneled inside and latched onto specific RNA molecules, labeling each with a specific barcode.
To amplify individual signals—each “point source”—the team used a common chemical reaction that rapidly amplifies DNA molecules, increasing their local concentration. DNA doesn’t like staying put inside the liquid interior of a cell, so the tags slowly begin to drift outwards, like a drop of dye expanding in a pool of water. Eventually, the DNA tags balloon into a molecular cloud that stems from their initial source on the biomolecule.
Be Part of the Future
Sign up to receive top stories about groundbreaking technologies and visionary thinkers from SingularityHub.


“Think of it as a radio tower broadcasting its signal,” the authors explained.
As DNA tag clouds from multiple points grow, eventually they’ll collide. This triggers a second reaction: two diffusing DNA molecules physically link up, spawning a unique DNA label that journals their date. This clever hack allows researchers to eventually triangulate the location of the initial sources: the closer the two points are in the beginning, the more labels they’ll form. The further apart, the fewer. The idea is very similar to cell phone companies using radio towers to track their users’ locations, in which they measure where signals from three or more towers intersect.
The cells are subsequently collected and their DNA extracted and sequenced—something that takes roughly 30 hours. The data is then decoded using a custom algorithm to transform raw data into gorgeous images. This is seriously nuts: the algorithm has absolutely no clue what a “cell” is, but still identified individual cells at their location inside the sample.
“The first time I saw a DNA microscopy image, it blew me away,” said Regev.
As proof of concept, the team used the technique to track cells that encode either green or red fluorescent proteins. Without any previous knowledge of their distribution, the DNA microscope efficiently parsed the two cell types, although the final images were blurrier than that obtained with a light microscope. The tech could also reliably map the location of individual human cancer cells by tagging the cells’ internal RNA molecules, although it couldn’t parse out fine details inside the cell.
A Whole New World
Thanks to DNA’s “stickiness,” the technique can be used to label multiple types of biomolecules, allowing researchers to track the location of and identify antibodies or surface proteins on any given cell type, although the team will have to further prove its effectiveness in tissue samples.
Although the resolution of DNA microscopy is currently on par, if not slightly lower than, that of a light microscope, it provides an entirely new perspective of biomolecules inside cells.
“We’ve used DNA in a way that’s mathematically similar to photons in light microscopy. This allows us to visualize biology as cells see it and not as the human eye does,” said Weinstein.
DNA microscopy already does things a light microscope can’t. It can parse out cells that visually look similar but have different genetic profiles, for example, which comes in handy for identifying various types of cancer and immune cells. Another example is neuroscience: as our brains develop, various cells drastically alter their genetic profiles while migrating long distances—DNA microscopy could allow researchers to precisely track their movements, potentially uncovering new ways to boost neuroregeneration or plasticity.
Only time will tell if DNA microscopy will reveal “previously inaccessible layers of information,” as the team hopes. But they believe that their invention will spark new ideas and uses in the scientific community.
“It’s not just a new technique, it’s a way of doing things that we haven’t ever considered doing before,” said Regev.
Image Credit: J. Weinstein et al./Cell 2019.
Dr. Shelly Xuelai Fan is a neuroscientist-turned-science-writer. She's fascinated with research about the brain, AI, longevity, biotech, and especially their intersection. As a digital nomad, she enjoys exploring new cultures, local foods, and the great outdoors.
Related Articles
Souped-Up CRISPR Gene Editor Replicates and Spreads Like a Virus

Your Genes Determine How Long You’ll Live Far More Than Previously Thought
Google DeepMind AI Decodes the Genome a Million ‘Letters’ at a Time
What we’re reading