Illumina, the biggest maker of genomic sequencing machines, say they’ve broken the “sound barrier” of sequencing. Their latest machine can transcribe 18,000 human genomes a year for $1,000 per genome—a mark dreamed of for over a decade.
When the human genome’s three billion molecular pairs were first fully transcribed (or sequenced) in 2003, it was deemed a seminal accomplishment, but one seemingly destined to be repeated only rarely. The project’s cost totaled $2.7 billion, and the first genomes were hundreds of millions of dollars apiece.
It was around that time that the $1,000 genome was first targeted. Why $1,000? Well, it’s a nice, round, pretty much arbitrary number—a sort of mile marker set by researchers. If ever it gets that cheap, scientists thought, we’ll be able to do anything.
In the early days, many experts thought it was an impossible target, Raymond McCauley, Chair of the Biotech Track at Singularity University, recently told Singularity Hub. But as costs began falling, the $1,000 genome started looking more realistic.
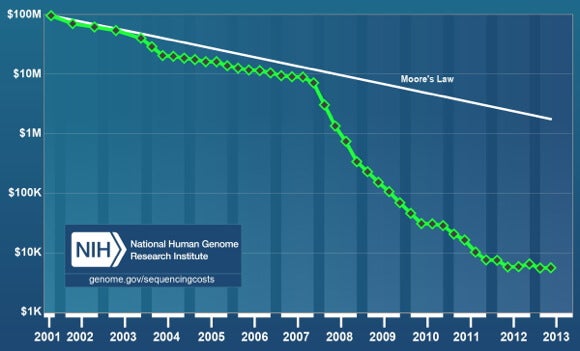
In 2006, the New York Times ran an article titled, “The Quest for the $1,000 Genome.” At the time, per genome sequencing costs had declined a full order of magnitude. Yet, despite a pace on par with Moore’s Law, if progress had continued along that line, the cost to sequence the human genome would still be over $1 million in 2014.
But it’s not. Not even close. Shortly after The New York Times article was published, next-generation sequencing hit the commercial market in a big way. What had, in the beginning, seemed an almost mythical goal started to look not only like a foregone conclusion—but one that would be realized far sooner than imagined.
Traditional sequencing (or Sanger sequencing) transcribes DNA using a complex, time-consuming, and costly chemical process. Next-generation machines, on the other hand, are more like supercomputers. They work in parallel, using brute force to simultaneously read millions of DNA base pairs at a time.
In the last six years, primarily on the back of next-generation sequencing technologies, costs have far outpaced Moore’s Law, falling four orders of magnitude from $10 million at the end of 2007 to under $5,000 at the end of 2013.
Now, owners of Illumina’s $10 million HighSeq X Ten may run 18,000 genomes a year for $1,000 a pop. That’s about a fifth of all the genomes that have been sequenced in history at a fraction of the expense.
It’s a staggering achievement a mere decade in the making. And as McCauley notes, “$1,000 is good base cost because it’s cheaper than an MRI; it’s cheaper than a lot of CT scans or a chest x-ray in some places.”
But it’s worth a moment’s pause to discuss what Illumina’s claim really means.
As the name implies, the HighSeq X Ten isn’t really one machine. It’s ten. The supercomputer analogy is instructive here too. Though there are more transistors per chip these days, the fastest computers mainly get faster by packing more chips into more cabinets—they still fill entire rooms.
Also, the cost of sequencing is a sensitive, much hyped subject. The $1,000 genome has been just around the corner for roughly two years now. Beyond flashy PR claims, cost calculations can be controversial.
In the past, firms would include the expense of chemicals but exclude key line items like depreciation and labor, sparking fierce debates online—a fact, no doubt, Illumina knows well. They were careful to include chemicals, depreciation, and labor (albeit cheap labor, according to some) in their estimate.

McCauley says, “You can tell that they’ve been working on it and waiting to get that marginal cost of chemicals and reagents down below a certain figure where the all-in cost for materials, the depreciation on the machine, and all of that, is right about at $1,000.”
Even so, he went on to tell us, the numbers haven’t been nailed down yet.
The National Human Genome Research Institute (NHGRI) publishes the most referenced graph (see above) on sequencing costs. NHGRI uses the cost of sequencing reported in research papers. That is, instead of going by a firm’s initial claim, they wait to see how the machine performs in practical use.
According to Illumina, they’ve sold machines to Korean genomics firm, Macrogen, the Broad Institute in Cambridge, Massachusetts, and the Garvan Institute of Medical Research in Sydney, Australia. The machines are due to ship the first quarter of 2014, and of course, they’ll take time to ramp up to full capacity.
There’s one more item worth noting—and this point may be somewhat lost in the chatter—you or I can’t go to a clinic to get our genome sequenced for $1,000. Not yet at least.
Machines like the HighSeq X Ten focus on sequencing for research purposes, for scientists conducting broad studies of thousands and tens of thousands of human genomes at a time. By comparing a large number of genomes, researchers can associate conditions and diseases with particular genes. This research is crucial if we are to replace DNA transcription with understanding and fluency in the language.
At $1,000 per genome, such work will likely accelerate.

McCauley notes that for a million dollars we could sequence one genome six years ago. Four years ago we could sequence ten; three years ago we could sequence a hundred; and now we can sequence a thousand.
“It’s one thing to say we’ve sequenced a single reference genome at the beginning of the century,” McCauley says, “It’s another thing to have a million genomes in a database. With this scale of improvement that becomes very reachable. And in fact, in aggregate, I think over the next couple years we are going to see a million genomes sequenced.”
But where all that data goes is an open question. A significant chunk may be tied up in proprietary, private pharmaceutical drug testing databases instead of being made publicly available by publicly-funded research groups. For the sake of faster progress, according to McCauley, the more of the latter we have, the better.
So, if practical results prove Illumina has truly claimed the $1,000 genome—what happens next? McCauley thinks next-generation sequencing is ripe to be supplanted by next-next generation sequencing. And today’s fast pace will largely continue.
“Anybody who enters into this, they’re living in dog years. They’ve got to figure out how to not only make the technology work but keep improving it at this pace of probably 4x or 5x a year—just to compete.”
And perhaps, increasingly, we’ll see a more diversified approach. Some work, for example, will focus more on speed or miniaturization.
Certain requirements, say sequencing an individual cancer genome, may prove more urgent and less concerned with ultra-low cost. Eventually, McCauley says, we’ll see handheld DNA scanners for environmental identification and security. James Bond stuff.
The current trajectory hints at such sci-fi devices, but not immediately.
Closer in, we need to get a handle on the impending flood of data. It doesn’t make sense to have a team of geneticists poring over every line. On the path to truly clinical genomics, expect software engineers and programmers to lay the next paving stones.
“Interpretation is king from now on,” McCauley says.
Images courtesy of Shutterstock.com




