Not Just Gene Editing—CRISPR Toolkit Expands With Trio of New Tricks

Share
CRISPR, the superhero of gene editing, just got a little more super.
In a trio of studies released last week in Science, leading CRISPR labs around the world unveiled some ultra-creative additions to the technique, transforming the hotshot editor into a virus detective or a sharp-eyed historian jotting down a cell’s entire history into DNA.
Like CRISPR, the new technologies are dubbed with cute acronyms such as CAMERA, DETECTR, and SHERLOCK. The sudden explosion of outside-the-box uses of CRISPR clearly shows that scientists are far from done exploring all the potential applications the technology has to offer.
To turn CRISPR into a true multi-tool, the teams got creative: CAMERA, the cell recorder, used rings of DNA as a readout for the cell’s history—say, exposure to antibiotics or toxins.
SHERLOCK, in contrast, added in “sacrificial” RNA molecules that are chopped up in the presence of virus or cancer DNA to create a positive signal. This shows up as a blue line on a paper strip, much like a pregnancy test.
“(It) highlights the really creative ways people are harnessing discoveries in CRISPR to build these synthetic pathways,” said Dr. Dave Savage, a protein engineer at the University of California, Berkeley, who was not involved in the studies.
The Cell Recorder
Airplane black boxes are a precious resource for investigators when a flight goes awry. Similarly, scientists have long hoped for a cellular time machine that details events in a cell’s life—doses of radiation, sprinklings of drugs, or bouts of internal turmoil that pushed a healthy cell towards a diseased state.
What better way to record a cell’s history than in DNA?
At Harvard University, Dr. David Liu took advantage of CRISPR’s accurate DNA-snipping ability and developed a “molecular historian” called CAMERA—short for CRISPR-mediated analogue multi-event recording apparatus (a mouthful!).
Here’s how it works. First, in bacteria cells, the team tweaked CRISPR’s guiding molecule—“guide” RNA—so that it only kicks into action after a trigger: say, an antibiotic or other chemical attack. Once active, the guide RNA beckons Cas9, the CRISPR scissors, to the target location. No guide RNA, no chopping action.
In addition to the modified CRISPR system, the team also gave cells two extra bits of DNA encoded in circular DNA called plasmids. Normally, the cell expresses the two plasmids at a steady ratio. But once the guide RNA is activated, it (and Cas9) goes after only one of the plasmids and chews it up—leaving the other intact.
In this way, the team can easily measure the ratio of the two plasmids. Using the system the team was able to detect if a cell was treated with tetracyclin, a common antibiotic.
But this first system only worked in bacterial cells. Liu and team further developed a second CAMERA that uses a modified Cas9. Rather than cutting a target gene, these scissors find a specific DNA letter and swap it with another.
As before, the system was triggered only in response to certain signals—drugs, nutrients, light, or even the cell’s signaling molecules, such as those related to cancer. The recorder not only logged the presence of a signal; by reading the fraction of swapped DNA letters, the team could also determine its duration and strength.
Unlike previous generation cell recorders, Liu’s system is highly sensitive, needing only a dozen cells to generate a strong signal. SCRIBE, a similar recorder developed by Dr. Timothy Lu at MIT back in 2014, required “orders of magnitude” more cells with a poorer signal-to-noise ratio.
CAMERA also boasts other useful features. It can, for example, record multiple signals simultaneously. The data can also be wiped using drugs that reset the plasmid ratio back to baseline.
Lu is impressed. The new work, he says, is “really beautiful stuff” and an “important advance.” Although any medical applications are a longs ways off, the system could help detect environmental pollutants or help scientists track the diverse sets of molecular signals that transform stem cells into neurons, muscles, or other cell types.
The Virus Hunters
Two additional studies explored ways to turn CRISPR into a sensitive one-step diagnostic tool.
DETECTR takes advantage of Cas9’s less famous cousin, Cas12.
Like Cas9, Cas12 follows a guide RNA towards a target and cuts it up.
Be Part of the Future
Sign up to receive top stories about groundbreaking technologies and visionary thinkers from SingularityHub.


But the scissors don’t stop there. A team led by Dr. Jennifer Doudna, a CRISPR pioneer at UC Berkeley, surprisingly found that Cas12 remains active after chopping up its intended target—it immediately hunts down single-stranded DNA molecules for destruction.
Tapping into this peculiar activity, the team added molecular neon signs to their guide RNAs, which begin to glow bright green once Cas12 activates.
In a proof-of-concept, they engineered several guide RNAs that bind to different strains of human papillomavirus (HPV), several of which cause cervical cancer. The DETECTR system was able to tease out two especially dangerous strains of HPV in a soup of different viral strains.
"This protein works as a robust tool to detect DNA from a variety of sources," says study author Janice Chen. "We want to push the limits of the technology, which is potentially applicable in any point-of-care diagnostic situation where there is a DNA component, including cancer and infectious disease."
The downside? Cas12’s over-zealousness could limit its use for treating genetic diseases in people. Editas, a CRISPR startup, has already licensed Cas12 for further development, and the finding could put a damper on their ambitions.
That said, it may be too early to throw out Cas12 as a gene editing treatment. While human DNA does unwind into single-stranded DNA (and thus a Cas12 target), the enzyme mostly binds to genomic DNA. This restricts its ability to roam around the cell hunting for potential single-stranded targets, explains Dr. Feng Zhang at the Broad Institute, who also published a CRISPR-based viral detection system last week dubbed SHERLOCK.
Based on a system first developed in 2017, SHERLOCK—Specific High Sensitivity Reporter unLOCKing—uses Cas13 as its cutting tool. Cas13 also has a wild streak, going into overdrive after it demolishes its original target.
With SHERLOCK, Zhang and colleagues added in “sacrificial” RNA molecules that create a signal once cleaved. In the presence of viral DNA or RNA—say Zika, Ebola, or dengue virus—Cas13 dices the viral target and the sacrificial RNA, thus releasing the positive signal.
SHERLOCK 2.0, published online last week in Science, is 100 times more sensitive than the original, and can detect up to four different targets at once.
It’s also highly practical for field use: all the reagents are bound to a paper strip, which is dipped into a test sample. If a line appears then the test is positive—no expensive specialty instruments required.
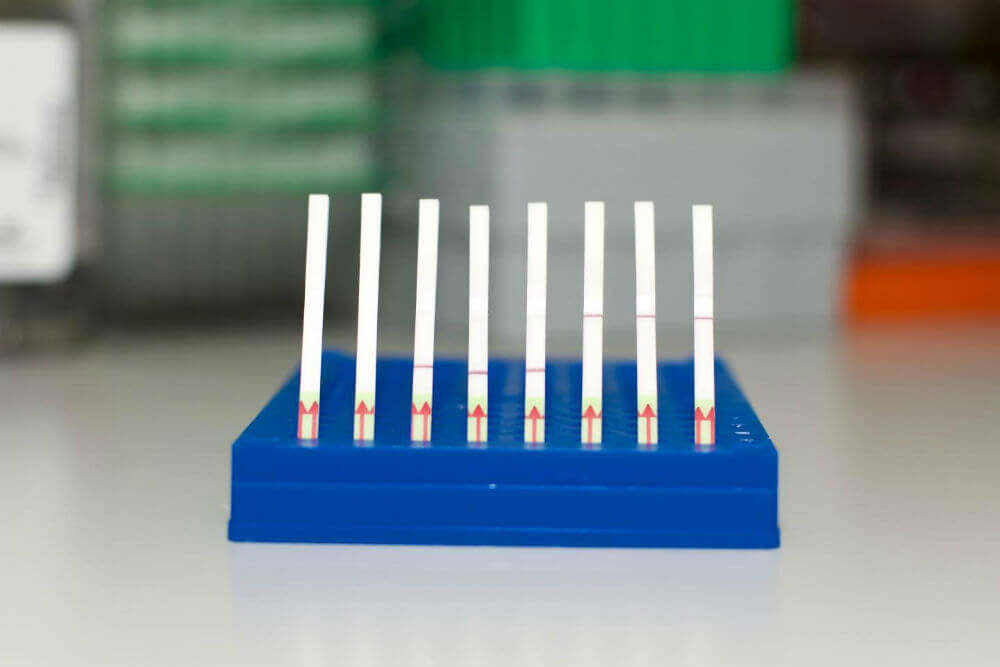
This is a collection of SHERLOCK paper test strips. (Left) Unused paper strips. (Middle) Paper tests displaying a negative SHERLOCK readout. (Right) Paper tests displaying a positive SHERLOCK readout. Image Credit: Zhang lab, Broad Institute of MIT and Harvard
Like DETECTR, this makes SHERLOCK especially useful during outbreaks. The tech can be easily modified to track other DNA molecules in the blood, for example, those normally associated with cancer or aging cells.
Together, the studies add to a growing research trend keen on exploring CRISPR applications beyond gene therapy. Compared to CRISPR-based therapeutics, which requires years of rigorous safety and efficacy testing, these “alternative” uses may quietly enter the scientific and diagnostic mainstream on a much faster timescale.
Although CRISPR’s new tricks remain to be tested in the field, experts are hopeful.
“The real power is what’s going to happen next,” said Savage. “Right now, the killer app is still to come.”
Image Credit: Anasta / Shutterstock.com
Dr. Shelly Xuelai Fan is a neuroscientist-turned-science-writer. She's fascinated with research about the brain, AI, longevity, biotech, and especially their intersection. As a digital nomad, she enjoys exploring new cultures, local foods, and the great outdoors.
Related Articles
In a First, Researchers Use Stem Cells and Surgery to Treat Spina Bifida in the Womb

Hackers Are Automating Cyberattacks With AI. Defenders Are Using It to Fight Back.
This Week’s Awesome Tech Stories From Around the Web (Through March 7)
What we’re reading