AI Discovers a New Class of Antibiotics After Scouring 12 Million Compounds
Share
Antibiotics have saved countless lives and are a crucial tool in modern medicine. But we're losing ground in our battle against bacteria. In the middle of the last century, scientists discovered whole new classes of antibiotics. Since then, the pace of discovery has slowed to a trickle, and the prevalence of antibiotic-resistant bacteria has grown.
There are likely antibiotics yet to be discovered, but the chemical universe is too big for anyone to search. In recent years, scientists have turned to AI. Machine learning algorithms can whittle enormous numbers of potential chemical configurations down to a handful of promising candidates for testing.
To date, scientists have used AI to find single compounds with antibiotic properties. But in a new study, published yesterday in Nature, MIT researchers say they've built and tested a system that can identify whole new classes of antibiotics and predict which are likely safe for people.
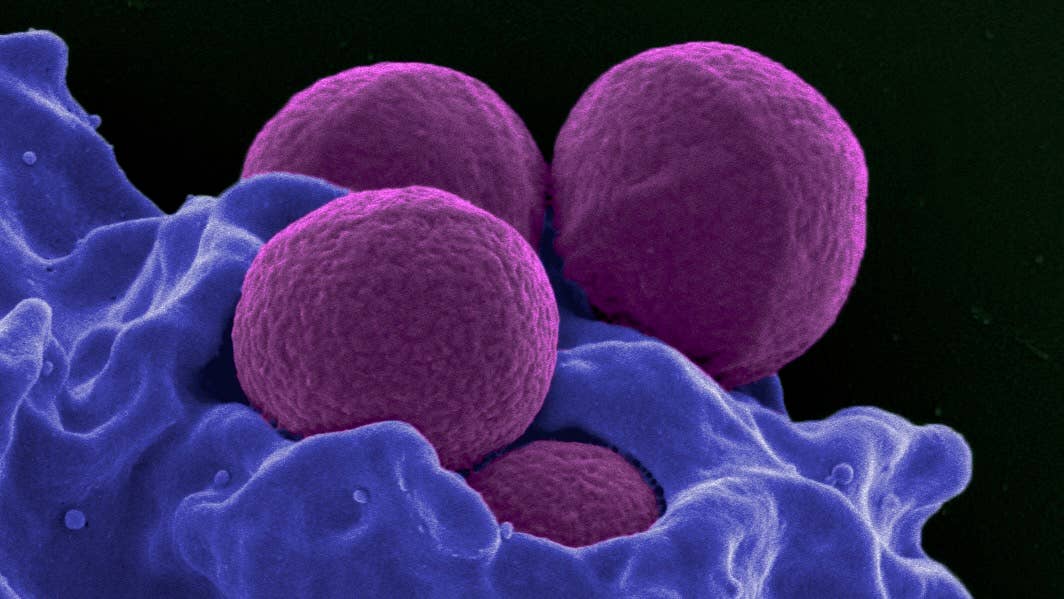
The AI sifted over 12 million compounds and found an undiscovered class of antibiotics that proved effective in mice against methicillin-resistant Staphylococcus aureus (MRSA), a deadly strain of drug-resistant bug.
While these AI-discovered antibiotics still need to prove themselves safe and effective in humans by passing the standard gauntlet of clinical testing, the team believes their work can speed discovery on the front end and, hopefully, increase our overall hit rate.
Exploring Drug Space
Scientists are increasingly using AI sidekicks to speed up the process of discovery. Most famous, perhaps, is DeepMind's AlphaFold, a machine learning program that can model the shapes of proteins, our body's basic building blocks. The idea is that AlphaFold and its descendants can speed up the arduous process of drug research. So strong is their conviction, DeepMind spun out a subsidiary in 2021, Isomorphic Labs, dedicated to doing just that.
Other AI approaches have also shown promise. An MIT group, in particular, has been focused on developing entirely new antibiotics to fight superbugs. Their first study, published in 2020, established the approach could work, when they found halicin, a previously undiscovered antibiotic that could readily take out drug-resistant E. coli.
In a followup earlier this year, the team took aim at Acinetobacter baumannii, "public enemy No. 1 for multidrug-resistant bacterial infections," according to McMaster University's Jonathan Stokes, a senior author on the study.
“Acinetobacter can survive on hospital doorknobs and equipment for long periods of time, and it can take up antibiotic resistance genes from its environment. It’s really common now to find A. baumannii isolates that are resistant to nearly every antibiotic,” Stokes said at the time.
After combing through 6,680 compounds in just two hours, the AI highlighted a few hundred promising candidates. The team tested 240 of these that were structurally different from existing antibiotics. They surfaced nine promising candidates, including one, abaucin, that was quite effective against A. baumannii.
Both studies showed the approach could work, but only yielded single candidates with no information on why they were effective. Machine learning algorithms are, notoriously, black boxes—what happens "between the ears" so to speak is often a complete mystery.
In the latest study, the group took aim at another known adversary, MRSA, only this time they chained several algorithms together to improve results and better illuminate the AI's reasoning.
Flipping the Switch
The team's latest antibiotic bloodhound trained on some 39,000 compounds, including their chemical structure and ability to kill MRSA. They also trained separate models to predict the toxicity of a given compound to human cells.
“You can represent basically any molecule as a chemical structure, and also you tell the model if that chemical structure is antibacterial or not,” Felix Wong, a postdoc at IMES and the Broad Institute of MIT and Harvard, told MIT News. “The model is trained on many examples like this. If you then give it any new molecule, a new arrangement of atoms and bonds, it can tell you a probability that that compound is predicted to be antibacterial.”
Once complete, the team fed over 12 million compounds into the system. The AI narrowed this enormous list down to around 3,600 compounds organized into five classes—based on their structures—it predicted would have some activity against MRSA and be minimally toxic to human cells. The team settled on a final list of 283 candidates for testing.
Be Part of the Future
Sign up to receive top stories about groundbreaking technologies and visionary thinkers from SingularityHub.


Of these, they found two from the same class—that is, they had similar structural components believed to contribute to antimicrobial activity—that were quite effective. In mice, the antibiotics fought both a skin infection and a systemic infection by taking out 90 percent of MRSA bacteria present.
Notably, while their previous work tackled Gram-negative bacteria by disrupting cell membranes, MRSA is Gram-positive and has thicker walls.
“We have pretty strong evidence that this new structural class is active against Gram-positive pathogens by selectively dissipating the proton motive force in bacteria,” Wong said. “The molecules are attacking bacterial cell membranes selectively, in a way that does not incur substantial damage in human cell membranes."
By making their AI explainable, the team hopes to zero in on structures that might inform future searches or contribute to the design of more effective antibiotics in the lab.
Final Exams
The key thing to note here is that although it appears the new antibiotics were effective in mice on a very small scale, there's a long way to go before you'd be prescribed one.
New drugs undergo rigorous testing and clinical trials, and many, even promising candidates, don't make it through to the other side. The field of AI-assisted drug discovery, more generally, is still in the early stages in this respect. The first AI-designed drugs are now in clinical trials, but none have yet been approved.
Still, the hope is to more quickly stock the pipeline with better candidates.
It can take three to six years to discover a new antibiotic suitable for clinical trials, according to the University of Pennsylvania’s César de la Fuente, whose lab is doing similar work. Then you have the trials themselves. With antibiotic resistance on the rise, we may not have that kind of time, not to mention the fact antibiotics don't have the return on investment other drugs do. Any help is welcome.
"Now, with machines, we’ve been able to accelerate [the timeline]," de la Fuente told Scientific American. "In my and my colleagues’ own work, for example, we can discover in a matter of hours thousands or hundreds of thousands of preclinical candidates instead of having to wait three to six years. I think AI in general has enabled that."
It's early yet, but if AI-discovered antibiotics prove themselves worthy in the coming years, perhaps we can maintain the upper hand in our long-standing battle against bacteria.
Jason is editorial director at SingularityHub. He researched and wrote about finance and economics before moving on to science and technology. He's curious about pretty much everything, but especially loves learning about and sharing big ideas and advances in artificial intelligence, computing, robotics, biotech, neuroscience, and space.
Related Articles
Souped-Up CRISPR Gene Editor Replicates and Spreads Like a Virus

Your Genes Determine How Long You’ll Live Far More Than Previously Thought
Google DeepMind AI Decodes the Genome a Million ‘Letters’ at a Time
What we’re reading
