
Scientists have worked out a non-invasive way to sequence a fetus’ genome, using only blood from the mother and saliva from the father-to-be. The technique could one day provide clinicians a much safer alternative than current methods which screen for genetic diseases. At the same time, the prospect of routine DNA sequencing of unborn babies has raised ethical concerns in many.
Since 1997 it has been known that pieces of fetal DNA circulate in the mother’s bloodstream during pregnancy. Both maternal and fetal DNA contribute to the “cell-free” DNA in the blood plasma, with maternal DNA accounting for 90 percent of the total and fetal DNA 10 percent. Dr. Jay Shendure and collegues at the Department of Genome Sciences at the University of Washington have taken advantage of the fetal DNA presence by sequencing it and then using the sequences of both the mother and father-to-be to predict the fetus’ entire genome to a 98 percent plus accuracy. We contacted Dr. Shendure and asked him about how his procedure might eventually change the future of prenatal genetic screens, and what work still remains to get there.
The obvious benefit of a genetic screen that requires only blood and saliva is that it is non-invasive. Today’s screens involve invasive procedures such as collecting fluid directly from the uterus (amniocentesis) or snipping off a piece of placental tissue (chorionic villus sampling). But in addition to being non-invasive, the new technique involves much more extensive sequencing – termed deep ultra-sequencing – than is normally performed for genetic screens. “The improved resolution is like going from being able to see that two books are stuck together to being able to notice one word misspelled on a page,” explained Jacob Kitzman in a news release from the University of Washington. Kitzman is a researcher in the lab of Dr. Shendure’s lab. He led the study with fellow researcher Matthew Snyder.
Sequencing from the baby’s umbilical cord following birth confirmed that their predicted sequence was 98.2 percent accurate. It also revealed that they’d correctly predicted 39 of the baby’s 44 de novo mutations not present in the mother’s genes.
The leap in resolution was made possible by a new technique and statistical modeling. Because the fetus’ DNA accounts for just 10 percent of total DNA in the mother’s blood the researchers needed to find a way to distinguish the two. A recently developed technique allowed them to identify haplotypes, groups of genes that are genetically linked on the same chromosome and thus inherited together. As each chromosome carries two copies of each gene, subtle genetic differences between each copy can be used to distinguish between haplotype pairs of a chromosome. When these haplotype pairs are measured in the mother’s blood they will, for the most part, be present in equal amounts. Because the fetus inherits one copy of a gene from the mother and one from the father, only one of the mother’s haplotypes will be found among the fetus’ DNA. So if one of the mother’s haplotypes is found to exist in greater proportion than its corresponding haplotype, one can conclude that it comes from the fetus. Using this method and comparing the father’s DNA sequence, the researchers were able to correctly identify which of the fetus’ genes with a 98.2 percent accuracy.

According to Dr. Shendure, it is the first time a fetus’ genome has been sequenced non invasively. The researchers then repeated the procedure and sequenced the genome of a fetus 8.2 weeks following conception, a more difficult task since there is less fetal DNA in the blood. Predictive power suffered as they accuracy dropped to about 92 percent.
The study was published June 6 in Science Translational Medicine.
Though the DNA from the parents is easy to acquire, the cost of sequencing and analysis make this procedure too costly to be of practical use in the clinic at the moment. But it’s a successful proof of concept and opens the door for improvements that would make it easier and less costly to perform. How long could it be before expectant parents are able to make their lives easier with a non-invasive screen? “These things are hard to predict,” Dr. Shendure said in an email, “but I would say at least five years. The remaining technical issues are fairly addressable, and the cost of sequencing is dropping on its own without us really having to do anything (i.e. we are simply riding this Moore’s Law). Sequencing costs probably need to drop another order of magnitude to make this work clinically, but…this seems not entirely unrealistic given that costs have dropped ~4 orders of magnitude in the last 7 or so years.”
He points out, however, that even as sequencing costs drop, their haplotype strategy would remain a challenge to feasibility. “One key advance that we still need to make technically is to figure out how to make the methodology for ‘haplotype-resolved’ genome sequencing scalable, faster, and cheaper. The way we did it in the current study is not terribly scalable and probably not fast enough. However, I think this goal can be achieved with a modest amount of additional work from our group and others working in this area.”
And technical issues, Dr. Shendure points out, are just the more straightforward hurdle that needs to be overcome before the screen can be a feasible tool. “There is tremendous work that needs to be done regarding genome interpretation for many of these diseases and then it’s always a big task to get any new test into clinical use, but even more so when it is a complex technique such as this one.”
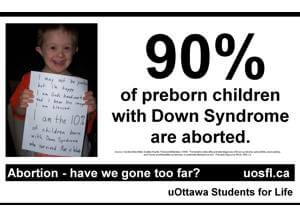
Still, the proof of concept study offers the possibility that prenatal genetic screens might be performed much more widely in the near future. Current screens only test for a small handful of genetic disorders such as cystic fibrosis and the blood disorder thalassemia. Screens for Down’s Syndrome are also common, and are sure to expand given we now have a test that detects the disorder from a blood sample. But there are more than 3000 single-gene (Mendelian) disorders the detection of which could benefit greatly from a user-friendly, non-invasive test. These Mendelian disorders are each rare on their own, but as a whole affect about 1 percent of births. A non-invasive screen would greatly expand detection of these disorders and allow doctors to modify prenatal care and lower the risk for complications during birth. More importantly, a non-invasive screen removes the risk of miscarriage associated with invasive amniocentesis.
As a non-invasive prenatal screen draws closer, the expectation that it will cause abortions to increase will undoubtedly be raised. But for many expectant mothers who want to screen but are kept away for fear of a miscarriage, swapping a 5-inch amniotic needle for a simple blood test, the screen would be a dream come true. Whether this procedure becomes a common fetal screening method within five years (given the drop in sequencing cost and increasingly more accurate and robust genome analyses), the potential benefits of complete fetal genome detection are immense and could have very broad ripple effects in healthcare and society as a whole.
[image credits: University of Washington, Scientific American, uOttawa Students For Life, and The Raw Story]
images: University of Washington, Scientific American, uOttawa Students For Life, and The Raw Story



