All of nature’s bewildering complexity can be reduced to a four letter code—DNA’s four bases: A, T, C, and G. But now scientists have shown the language at the heart of life is not set in stone by getting bacteria to build proteins that don’t exist in nature using two unnatural bases.
The blueprint for every organism is stored in its DNA, which holds the instructions for the proteins that act as the building blocks of life. These instructions consist of sequences of codons—three-letter strings of bases that encode for particular amino acids. Proteins are built from combinations of amino acids, and the sequence of codons determines what protein a particular string of DNA codes for.
There’s a temptation to assume that millions of years of evolution would yield the most efficient possible system for storing genetic information, but there’s actually a huge amount of redundancy. Multiple codons with different combinations of bases can code for the same amino acid, which is why 64 possible combinations only result in 20 amino acids.
The relatively limited palette nature has been using so far has prompted many researchers to investigate ways to expand the language at the heart of genetics. Some have looked at reassigning the meaning of words by repurposing redundant codons to code for new amino acids. Others have tried to expand the alphabet by modifying the bases at its heart or even creating entirely new ones.
Now, for the first time, researchers have demonstrated semi-synthetic bacteria that not only incorporate unnatural bases in their genetic code, but can use them to build proteins from amino acids that don’t exist in nature.
The addition of two new bases—made from the chemicals dNaM and dTPT3, but informally referred to as X and Y, respectively—theoretically allow an organism to code for up to 152 new amino acids, which the creators hope could become the basis for new protein-based medicines.
“I would not call this a new life form—but it’s the closest thing anyone has ever made,” Floyd Romesberg, a professor at the Scripps Research Institute who led the study, said in a press release. “This is the first time ever a cell has translated a protein using something other than G, C, A or T.”
Romesberg’s lab first demonstrated E. coli bacteria that could copy the unnatural X and Y bases back in 2014, but it was only earlier this year that they managed to get the bacteria to store them stably and reliably pass them onto their daughter cells as they divided.
Their latest work, reported in the journal Nature, is a significant step forward because they were able to get the bacteria to transcribe these bases into RNA that was used to build proteins incorporating an unnatural amino acid.
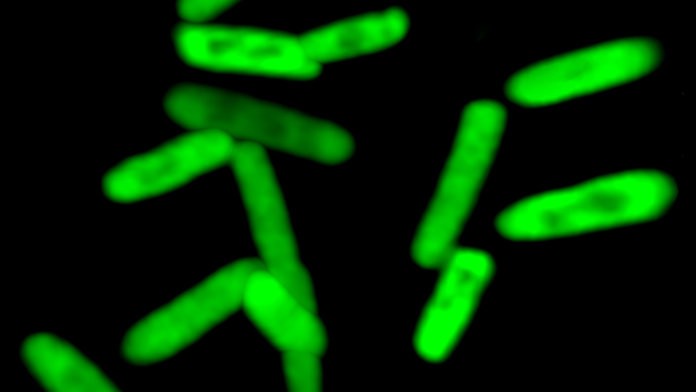
They used this ability to create a variant of green fluorescent protein with very high efficiency. While the protein was not functionally different from the original, the research demonstrates that the new base pairs are able to contribute to every stage of the process of retrieving information from the genetic code and using it to build proteins.
The main promise of this expansion of the genetic alphabet is the increased flexibility it could give to chemists and biologists trying to use living organisms to build new drugs, polymers, or catalysts. The most immediate promise is in medicine, and Romesberg has created biotech startup Synthorx to find commercial applications for his research.
Romesberg told Nature the company is trying to incorporate unnatural amino acids into a protein called interleukin-2 that regulates the activity of white blood cells. Synthetic versions of the protein are used as a cancer drug, but can have severe side effects. The addition of unnatural amino acids could potentially be used to rework the molecule to reduce toxicity, make it easier for cells to take up, or get it to break down faster.
Further down the line, rather than just using these semi-synthetic microbes as factories for unusual proteins, Romesberg told the Washington Post it may even be possible to use these alien bases to create organisms with capabilities beyond those of their natural cousins.
Despite the promise, though, the authors admit the method is still in its early days. Every X or Y had to be sandwiched between natural bases, which means it’s unlikely they’ll be able to replicate long stretches of unnatural bases.
But the main contribution of the research could be the shaking up of assumptions about both the essential ingredients for life and the potential recipes. Not only are X and Y unnatural bases, they don’t even link together using the hydrogen bonds natural bases rely on, which were considered an essential characteristic of DNA. Instead, they were designed to use hydrophobic forces to bind together and repel the other natural bases.
This highlights the flexibility of the rules governing DNA and suggests there is considerable scope for scientists to get creative with the code of life. As Romesberg told MIT Tech Review, “Life as we know it may not be the only solution, and may not be the best one.”
Image Credit: Bill Kiosses, The Scripps Research Institute