It used to be the stuff of science fiction: bringing a long-dead species back from extinction by painstakingly piecing together its full DNA sequence, or genome.
It’s not quite as straightforward as Jurassic Park would have us believe, but in the age of DNA editing, the idea of cloning an extinct species is no longer purely the realm of fantasy.
Our team at the DNA Zoo has hopefully taken a step towards creating a blueprint to clone one of Australia’s most loved, and most missed, extinct species: the thylacine, or Tasmanian tiger.
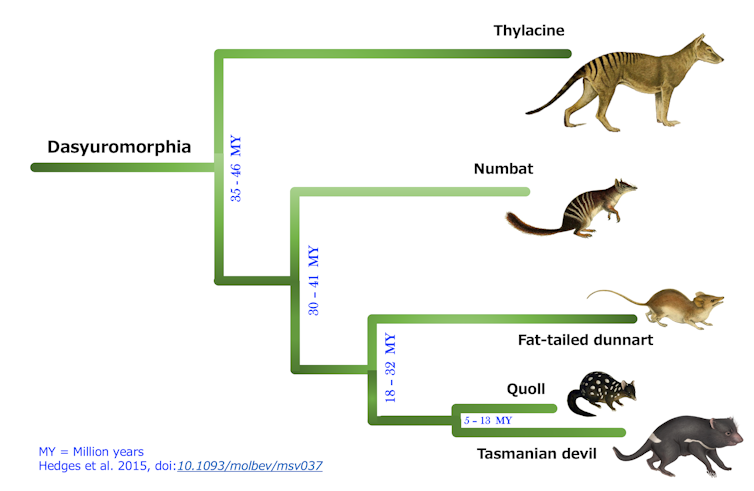
We’ve done it not by studying the thylacine itself, but by completing a chromosome-length 3D genome map of one of its closest living relatives: the numbat.
The striped, termite-eating numbat is Western Australia’s faunal emblem, and now lives only in small pockets of that state, although it once roamed throughout southern Australia. Crucially, numbats and thylacines shared a common ancestor that lived some time between 35 million and 41 million years ago—relatively recently in evolutionary terms.
Both these enigmatic creatures have stripes, but that’s not where the similarity ends—as much as 95% of their DNA may be identical.
Decoding the full numbat genome therefore raises the tantalizing prospect of being able to piece together the thylacine’s genetic sequence, which in turn would offer the tantalizing prospect of reintroducing one of Australia’s most iconic lost species.
No doubt this will be more challenging than the famous bid to resurrect the woolly mammoth using DNA from the Asian elephant. But the release of the numbat genome makes the thylacine’s resurrection a more realistic prospect than ever before.
The numbat is the latest marsupial genome sequence from this family compiled by our team at the DNA Zoo, following on from the Tasmanian devil, quoll, and dunnart. We’ve acquired samples of more than 500 mammals from around the world, and aim to make all their genomes available for conservation and open-access research.
We are also working on a detailed genomic analysis of most Australian carnivorous marsupials, and will ultimately produce a full peer-reviewed publication in a journal. But now, by sharing the sequence publicly at this stage of our research, we can offer a valuable resource to other scientists and conservationists studying numbats and other marsupials. Given the conservation threats they face, time is ticking fast.
Genes From Thylacines
The first draft of the Tasmanian tiger genome was pieced together in 2018, using century-old museum samples. But this version is very fragmentary—several key gaps still need to be filled to piece this puzzle together into a comprehensive genome sequence. Unfortunately, the old museum samples didn’t provide enough high-quality DNA to resolve these issues.
So how do you reconstruct something without some seemingly essential ingredients? This is where the genome of the thylacine’s closest living cousin—the numbat—can help. Our new high-resolution numbat genome map can help us fill in the missing bits of the thylacine genome.
There will still be significant hurdles between having a complete thylacine genome and cloning a thylacine for real. But what takes this scenario from science fiction to potential reality is CRISPR gene-editing technology—a set of enzymes that allow scientists to target very particular snippets of DNA.
CRISPR has been referred to as a kind of “molecular scissors” that allow the precise selection and insertion of DNA from specimens, making the “de-extinction” of the thylacine or other species a realistic prospect by allowing geneticists to selectively “repair” the missing bits of its genome.
With the help of this and other synthetic biology tools, geneticists could conceivably piece together a set of chromosomes that could then be inserted into an egg cell with its existing nucleus removed, allowing the new DNA to act as the egg’s genetic blueprint. This is the technique being pursued by a US research group aiming to clone the mammoth by using the DNA of its closest living relative, the Asian elephant, to fill in the missing bits of mammoth DNA.
Science Fiction or Science Future?
Around the world, rapid advancements in embryology and genetics are opening up the possibility of resurrecting extinct species—or at least creating something that’s close enough to the original that it will develop and grow properly.
In 1996, British scientists successfully cloned a sheep, called Dolly. Then, in 2017, Chinese researchers used the same technique to create two genetically identical long-tailed macaques.
Through the growing field of synthetic biology and precise genome-editing technologies such as CRISPR, Harvard geneticist George Church has launched Colossal, a biotech company that has initially set on creating an elephant-mammoth hybrid, with the first calves expected in six years.
Helping Numbats First
Of course, the numbat is one of Australia’s most loved native marsupials in its own right.
Like the Tasmanian Tiger, it too was on the verge of extinction during the late 20th century, but extensive conservation efforts as well as government and community intervention are helping its numbers gradually bounce back.
Still, with fewer than 1,000 numbats left in the wild and the species still officially listed as endangered, our genetic blueprint hopefully paves the way for better numbat conservation information for our scientists on the front line. Many of these scientists are fighting the very genetic diseases threatening to exterminate numbats.
There is a still a long road ahead before the thylacine could be cloned. But if it works, the end goal of any de-extinction effort surely is to reintroduce animals to the wild.
If that were to happen, the thylacine already has one advantage over many de-extinction candidates: appropriate habitat. With reserves covering about half of Tasmania today, there would be ample places for thylacines to live, still teeming with the prey animals they used to eat.
There is no question it could be reintroduced to the Tasmanian bush. There is also good reason to do so: the thylacine was Tasmania’s key carnivore. Putting it back atop the food chain could help restabilize ecosystems that are under threat.
If and when that dream becomes reality, thylacines would owe a debt of gratitude to their little cousin, the humble numbat.![]()
This article is republished from The Conversation under a Creative Commons license. Read the original article.



